
This site is a fork of the original PRABI NPS@ server
|
[HOME]
[DESCRIPTION]
[HELP]
[NEWS]
[CONTACT]
[Geno3D]
July 22, 2024: NPS@ updated (see NEWS).
July 25 17h00 (Paris Time) - July 26 12h00 (Paris Time): NPS@ service interruption.
MAFFT help
 A brief introduction to MAFFT
A brief introduction to MAFFT
MAFFT identifies homologous regions rapidly by means of the fast Fourier transform (FFT).
MAFFT is classified as a similarity-based method and takes evolutionary information into account.
MAFFT 7 has several new features, including options for adding unaligned sequences into an existing alignment, adjustment of direction in nucleotide alignment, constrained alignment and parallel processing, which were implemented after the previous major update.
 Availability in NPS@
Availability in NPS@
MAFFT is available :
The set to align cannot have more than 1,000,000 characters.
So, for example, you can use MAFFT to align a set of similar sequences built from a BLAST search.
 Parameters
Parameters
Different alignment strategies are available, as well as sequence output order and output tree file generation.
The output format is CLUSTAL W.
 NPS@ MAFFT output example
NPS@ MAFFT output example
The NPS@ MAFFT output is divided into three parts.
-
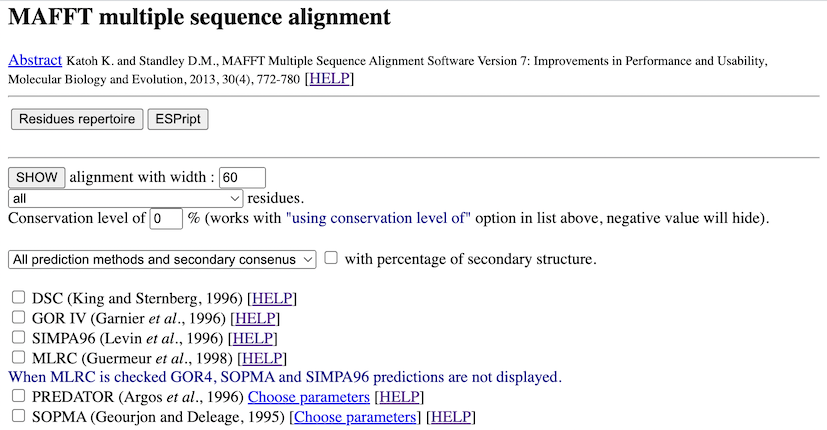
PART 1:
After the reference, you have buttons to post-process the multiple alignment with other software like HMMBUILD, Repertoire, ...
In this part, you have also a form to work with the alignment. Your choices are validate when you click on the SHOW button. You can :
- show residues with different options (identical(*), strongly similar (:), weakly similar (.), ...). The most
interesting option is the "using conservation level" one. With it you, display only residues that have a conservation level
equal or above the value you type in.
- select secondary structure prediction methods to compute and display. This in the case of proteins and when you have no
more than 50 sequences. A method with the "(Do it alone)" sentence has to be computed alone (it must be
the only one newly selected before the next click on "SHOW" button). Otherwise, you won't have your response because of the
timeout (these methods can take 5 minutes or more by sequence). You can also show all methods with or without
secondary consensus or only the consensus. You can
also see the percentage of each secondary structure element (HETC...) for each method.
-
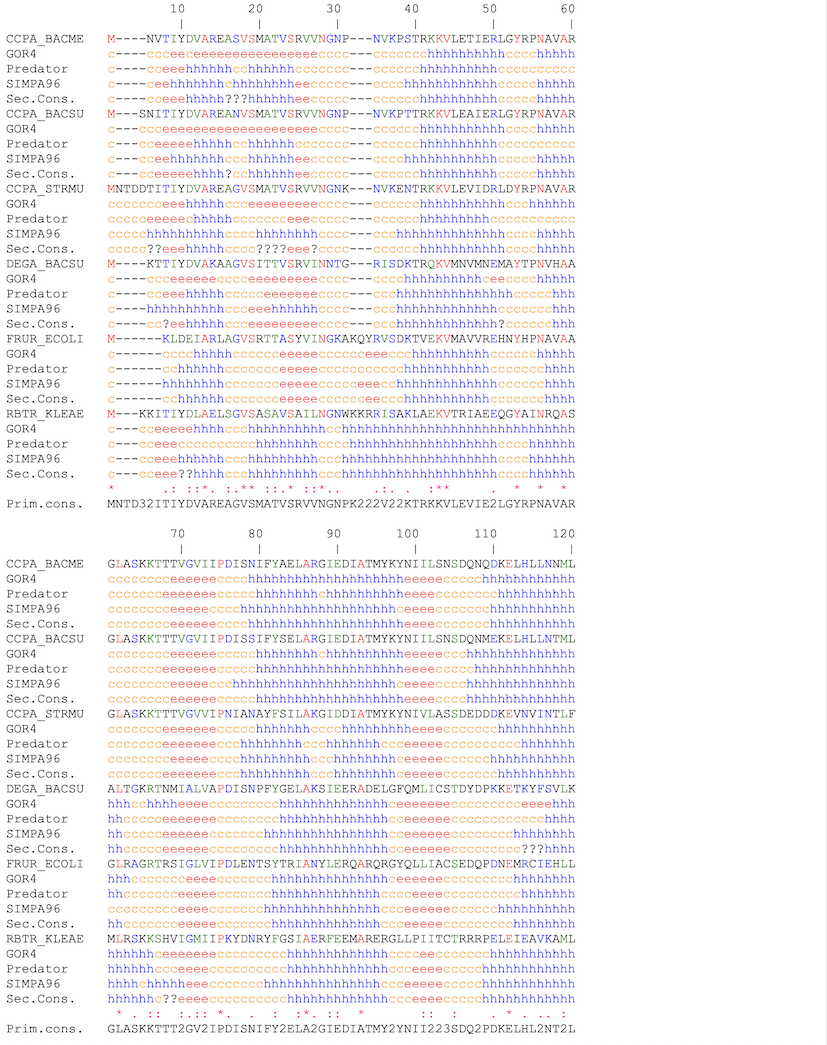
PART 2:
It's the color coded alignment with or without secondary structure predictions inserted.
-
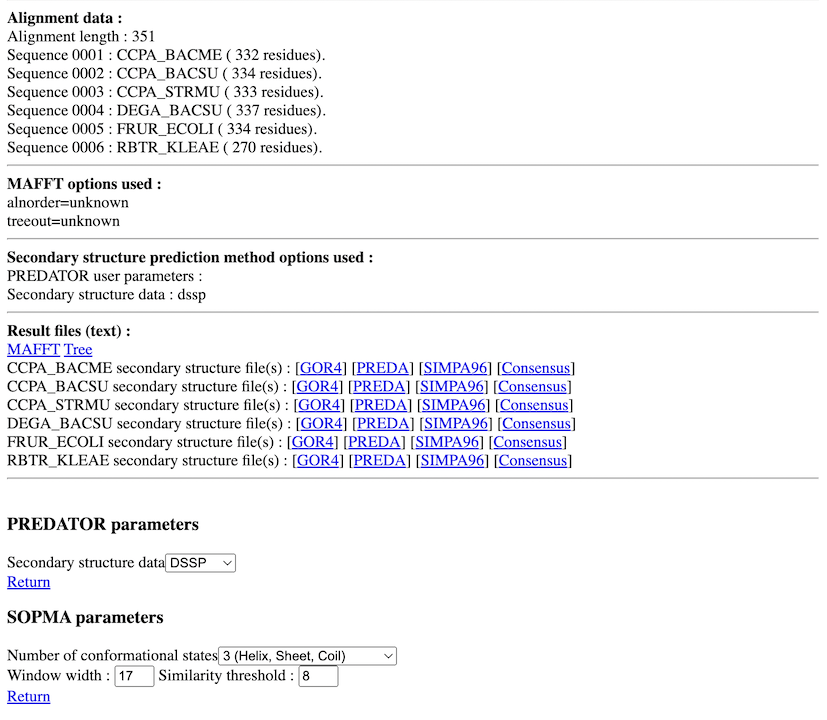
PART 3:
You have :
- The percentage of each secondary structure element (if wanted) for each prediction and for each sequence.
- Some data on the alignment (length, number of identities,...).
- MAFFT options used.
- Links on result text files (MAFFT, secondary structure prediction method outputs,...).
- Secondary structure prediction method parameters choice.
 References
References
Last modification time : Fri Dec 8 17:22:09 2023. Current time : Sat Jul 27 10:02:37 2024. User : public@3.149.244.45.


 A brief introduction to MAFFT
A brief introduction to MAFFT Availability in NPS@
Availability in NPS@ Parameters
Parameters NPS@ MAFFT output example
NPS@ MAFFT output example

