
This site is a fork of the original PRABI NPS@ server
 This site is a fork of the original PRABI NPS@ server |
[HOME] [DESCRIPTION] [HELP] [NEWS] [CONTACT] [Geno3D]
July 22, 2024: NPS@ updated (see NEWS).
July 25 17h00 (Paris Time) - July 26 12h00 (Paris Time): NPS@ service interruption.
 A brief introduction to SOPMA
A brief introduction to SOPMA
SOPMA (Self-Optimized Prediction Method with Alignment) is an improvement of SOPM method. These methods are based on the homologue method
of Levin et al.. The improvement takes place in the fact that SOPMA uses information from a multiple sequence alignment (MSA)
of protein belonging to the same family. If there are no homologous sequences the SOPMA prediction is the SOPM one.
Warning : It can be long to compute. So, be careful when using it in alignment (or it will exceed the NPS@'s maximum computing time).
NPS@ is the original server for this method.
 Availability in NPS@
Availability in NPS@
This method is available :
 Parameters
Parameters
You can set the number of conformational states to predict : 3 or 4.
The window size parameter sets the length of the peptides to use.
The similarity threshold parameter is the threshold below which a subject peptide is rejected when it's compared with a query peptide of the sequence.
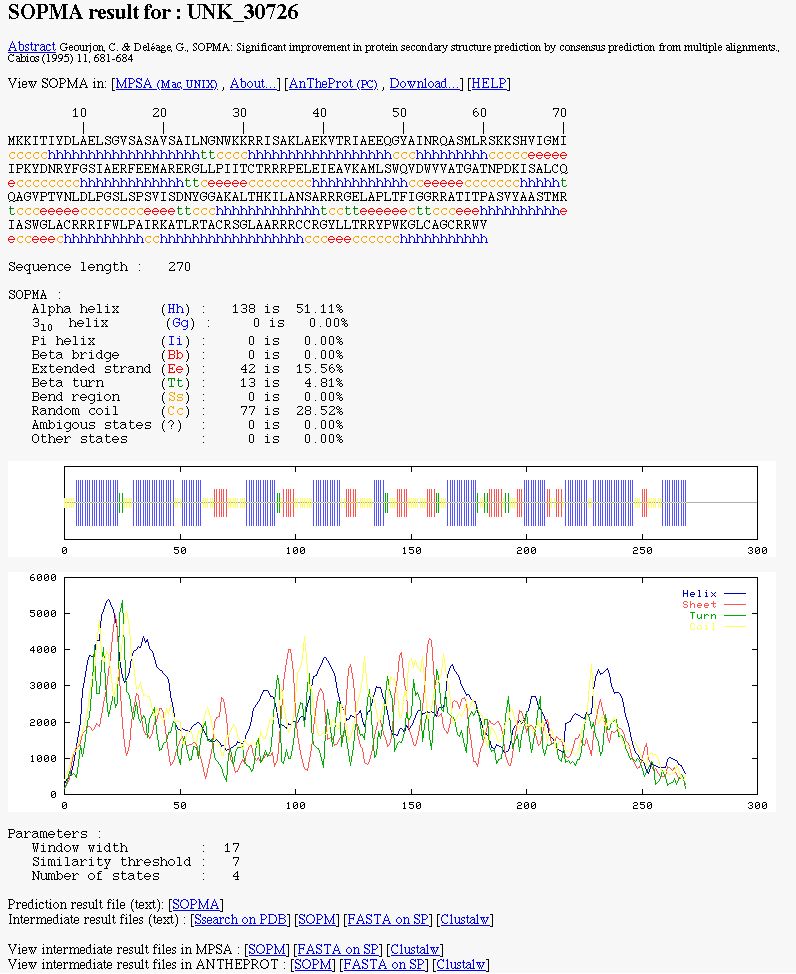
 NPS@ SOPMA output example
NPS@ SOPMA output example
You can see:
| © 1998-2024 |
|
|
|
Legal notice |