
This site is a fork of the original PRABI NPS@ server
|
[HOME]
[DESCRIPTION]
[HELP]
[NEWS]
[CONTACT]
[Geno3D]
July 22, 2024: NPS@ updated (see NEWS).
July 25 17h00 (Paris Time) - July 26 12h00 (Paris Time): NPS@ service interruption.
PSI-BLAST help
 A brief introduction to PSI-BLAST
A brief introduction to PSI-BLAST
The BLAST algorithm could use a position-specific matrix (profile) in place of a query sequence and associated substitution matrix. The
PSI-BLAST (Position-Specific Iterated BLAST) is an automated procedure generating the matrix from BLAST result and a modified BLAST
algorithm to take as input this matrix. The PSI-BLAST is an iterative process with a first step being a standard BLAST.
For further details, see the NCBI PSI-BLAST tutorial.
 Availability in NPS@
Availability in NPS@
PSI-BLAST is available :
 Parameters
Parameters
Some parameters are currently not available for the user.
By default the number of description and the number of alignment are set to 1000.
The expected threshold is set to 10.0.
The comparison matrix is BLOSUM62.
The default behavior is selected for gap opening (11) and gap extending (1) costs.
The expectation value threshold for inclusion in multipass model is 0.002.
The maximum number of passes is 7.
To retrieve these informations see the line "Information and statistics : [PSI-BLAST]" in
NPS@ PSI-BLAST result file.
 NPS@ PSI-BLAST output example
NPS@ PSI-BLAST output example
The NPS@ PSI-BLAST output is divided into three parts.
-
PART 1:
In this part, you have :
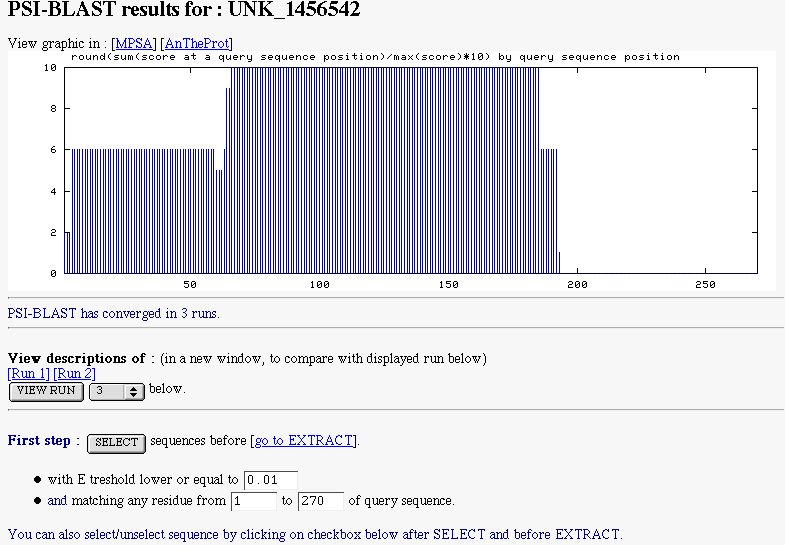
- a graphic indicating the similarity found along the query sequence, which is computed from PSI-BLAST PSSM-sequence alignments
- a line to say if PSI-BLAST has converged or not
- a form to select subject sequences. You can select sequences by indicated the maximal E threshold to do it (by default
NPS@ sets it to expectation value threshold for inclusion in multipass model). You can also select
subject sequences matching a particular region of the query
-
PART 2:
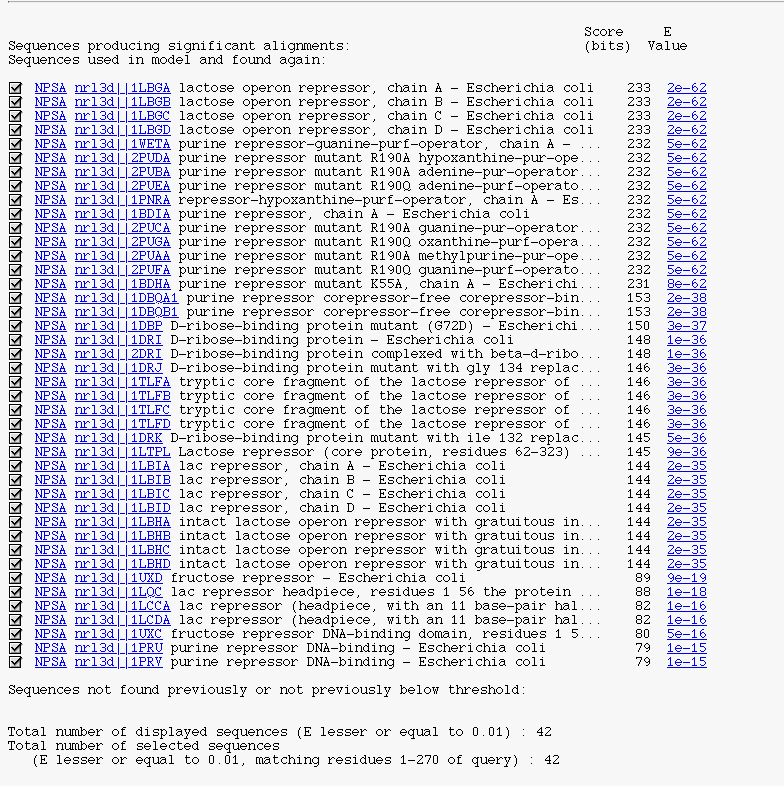
It's the PSI-BLAST description block in a 'HTMLized' form.
You can see :
- a checkbox to select/unselect subject sequence.
- the NPSA link allows you to apply NPS@ methods on the corresponding sequence after it is extracted from the query database
- the database link to retrieve the database entry
- the alignment link (on expected value) to see PSI-BLAST alignment between the query sequence and the current subject sequence
- the number of displayed sequences
- the number of selected sequences
-
PART 3:
You have :
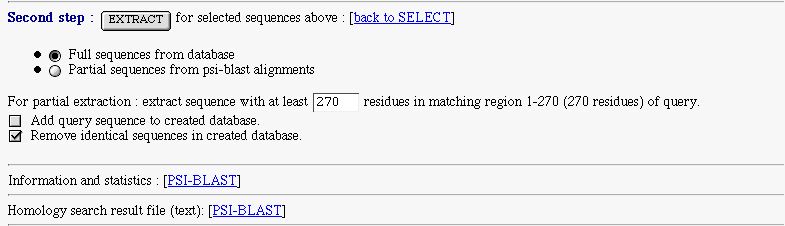
- an extract form to extract selected sequence and make a database. You can extract full or partial sequences.
The full extract is made from the database.
The partial extract, is made for subject sequence that match the query one in the indicated region. The extracted sequence is retrieved from the PSI-BLAST alignment. You can set the minimal length of the extracted sequence
- a checkbox to add your query sequence in the created database
- a checkbox to remove identical sequences in the created database
- a link on PSI-BLAST information and statistics
- a link on the original PSI-BLAST result text file
 References
References
Last modification time : Mon Dec 5 15:44:55 2022. Current time : Sat Jul 27 10:22:00 2024. User : public@3.12.120.25.


 A brief introduction to PSI-BLAST
A brief introduction to PSI-BLAST Availability in NPS@
Availability in NPS@ Parameters
Parameters NPS@ PSI-BLAST output example
NPS@ PSI-BLAST output example

