
This site is a fork of the original PRABI NPS@ server
|
[HOME]
[DESCRIPTION]
[HELP]
[NEWS]
[CONTACT]
[Geno3D]
July 22, 2024: NPS@ updated (see NEWS).
July 25 17h00 (Paris Time) - July 26 12h00 (Paris Time): NPS@ service interruption.
Analyse a set of sequences
 In NPS@ you can work with a set of sequences.
In NPS@ you can work with a set of sequences.
The set can be :
Look here for supported format (i.e. Pearson/FASTA) in NPS@.
 What can you do then ?
What can you do then ?
You can :
- align its sequences with KALIGN, MAFFT or MUSCLE (depending on the data size limit of 1,000,000 characters)
- run a sequence similarity search with BLAST or FASTA or PSI-BLAST or SSEARCH, the set will be the searched one
- apply NPS@ methods on each sequence of the set
- edit the set by selecting/unselecting sequence and writing a new set
- count the number of sequence in it
- see it as a text file
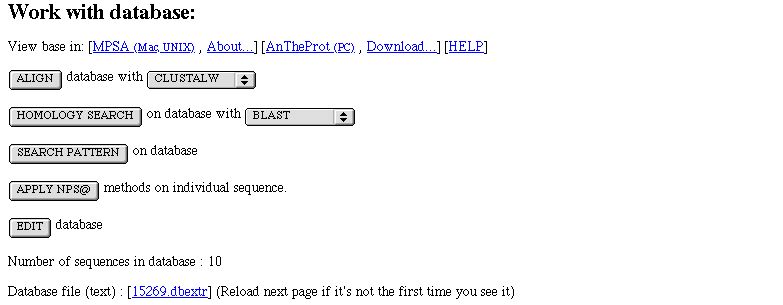
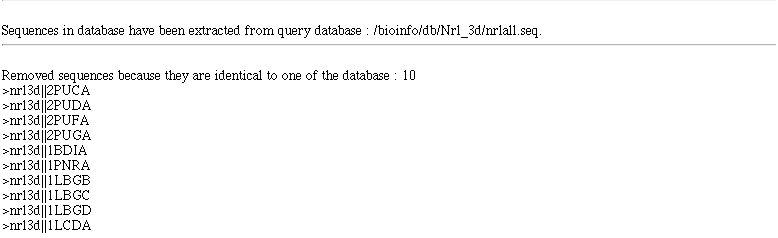
 When you come from an extract form (e.g. after BLAST) few more informations are displayed.
When you come from an extract form (e.g. after BLAST) few more informations are displayed.
- the database on which full sequence extraction has been made or it says you that it come from a partial extraction
- removed sequence identifiers in built set (when this option has been selected)
Last modification time : Wed Feb 15 16:45:35 2023. Current time : Sat Jul 27 08:25:02 2024. User : public@3.133.138.31.


 In NPS@ you can work with a set of sequences.
In NPS@ you can work with a set of sequences. What can you do then ?
What can you do then ?
 When you come from an extract form (e.g. after BLAST) few more informations are displayed.
When you come from an extract form (e.g. after BLAST) few more informations are displayed.