
This site is a fork of the original PRABI NPS@ server
|
[HOME]
[DESCRIPTION]
[HELP]
[NEWS]
[CONTACT]
[Geno3D]
July 22, 2024: NPS@ updated (see NEWS).
July 25 17h00 (Paris Time) - July 26 12h00 (Paris Time): NPS@ service interruption.
The NPS@ Web server
 NPS@ stands for Network Protein Sequence Analysis (NPSA altered in NPS@ to refer to its access by the Internet network).
NPS@ stands for Network Protein Sequence Analysis (NPSA altered in NPS@ to refer to its access by the Internet network).
 NPS@ is an interactive Web server dedicated to protein sequence analysis available for the biologist community.
NPS@ is an interactive Web server dedicated to protein sequence analysis available for the biologist community.
 NPS@ is the little web "brother" of ANTHEPROT and MPSA software. The guiding philosophy is the same for these two software and for this "webware". We hope they are useful and user-friendly.
NPS@ is the little web "brother" of ANTHEPROT and MPSA software. The guiding philosophy is the same for these two software and for this "webware". We hope they are useful and user-friendly.
 NPS@ is one service of the "Pôle Rhône-Alpes de Bio-Informatique" (PRABI).
NPS@ is one service of the "Pôle Rhône-Alpes de Bio-Informatique" (PRABI).
 NPS@'s history
NPS@'s history
- December 18th, 2019 : this NPS@ fork opening at https://npsa.lyon.inserm.fr by Christophe Combet.
- February 17th, 2003 : NPS@ version 3 is available. This third version was written by Christophe Combet, Céline Charavay, Christophe Geourjon, Christophe Blanchet and Gilbert Deléage.
- April 2nd, 1999 : NPS@ version 2 is available. This second version was written by Christophe Combet, Christophe Geourjon, Christophe Blanchet and Gilbert Deléage.
- April 9th, 1998 : NPS@ version 1 is available. This first version was written by Gilbert Deléage, Christophe Blanchet, Christophe Combet and Christophe Geourjon.
 What kind of analysis can you carry out with NPS@ ?
What kind of analysis can you carry out with NPS@ ?
- Homology search by means of pairwise sequence alignment or position-specific scoring matrix / profile:
FASTA,
BLAST,
PSI-BLAST
and SSEARCH
on sequence databases such as
European Nucleotide Archive (ENA),
UniProt Knowledge Base (UniProtrKB, Swiss-Prot),
Protein Data Bank (PDB),
AlphaFold Database or
ESM Metagenomic Atlas
.
- Multiple sequence alignment with
KALIGN,
MAFFT,
MUSCLE
.
- Protein secondary structure prediction including a consensus prediction of those methods. Available methods are
DSC,
GORIV,
MLRC,
PREDATOR,
SIMPA96,
SOPMA
.
- Primary structure analysis such as :
Coil-coiled domains prediction,
Coloring of amino-acid residues,
Composition in amino-acid residue,
Helix-Turn-Helix (HTH) DNA-binding motif prediction and
PROSITE motifs / patterns / signatures scan (PROSCAN) for protein domains, families and functional sites annotation
.
 What do you need to use NPS@ ?
What do you need to use NPS@ ?
- one sequence (see help).
- a sequence file in Pearson/FASTA format (see help).
 What are NPS@'s strong points ?
What are NPS@'s strong points ?
- All methods proposed by NPS@ are piped. The output of one method could be the input of another one. For example, after you've performed a sequence similarity search (e.g. BLAST), you can make a database of full or partial sequences. Thereafter, these sequences could be aligned by multiple sequence alignment programs (e.g. KALIGN, MAFFT, MUSCLE), or you can apply NPS@'s methods on each sequence of the database. And this, with no cut and paste, as data transfer are done on server side and format conversion are ensured when needed.
- You can insert protein secondary structure prediction in multiple sequence alignment.
- You can upload your own sequence file and apply NPS@'s methods on it.
- You can download NPSA's data in protein sequence analysis softwares on your local computer for further analysis, to save them or insert them in an article.
- The NPSA link allows you to apply NPS@'s methods on a sequence. Even more, when the sequence comes from a 3D database (PDB), you have some useful links to retrieve and work with 3D data.
- NPS@ offers links on international databases (e.g. ENA, UniProtKB, Swiss-Prot, PDB).
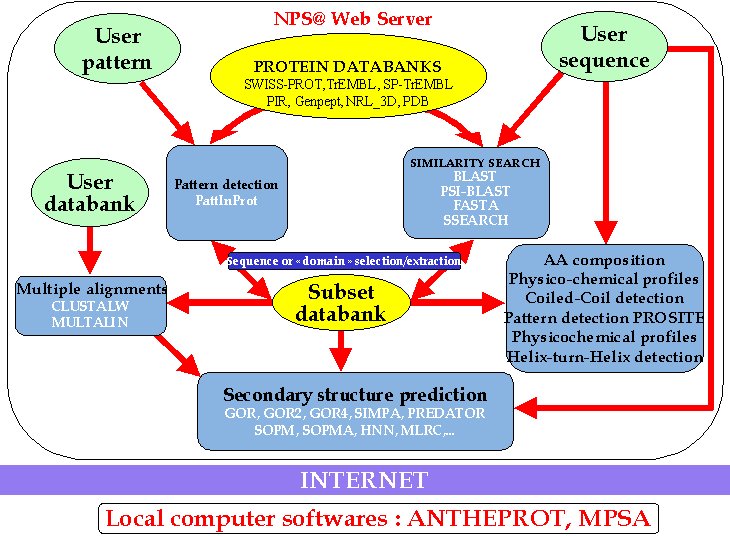
 NPS@'s flow chart.
NPS@'s flow chart.
 NPS@'s help index page.
NPS@'s help index page.
 To cite NPS@:
To cite NPS@:
NPS@: Network Protein Sequence Analysis
TIBS 2000 March Vol. 25, No 3 [291]:147-150
Combet C., Blanchet C., Geourjon C. and Deléage G.
We will be glad to receive a reprint of article citing NPS@.
This will be a contribution and an encouragement in developing this server.
 NPS@'s limitations:
NPS@'s limitations:
The CPU time can't exceed 30 minutes for one computation. For longer job, you can contact us.
 View number of jobs performed by NPS@.
View number of jobs performed by NPS@.
Last modification time : Mon Feb 5 16:42:42 2024. Current time : Sat Jul 27 08:29:44 2024. User : public@3.149.214.144.


 NPS@ stands for Network Protein Sequence Analysis (NPSA altered in NPS@ to refer to its access by the Internet network).
NPS@ stands for Network Protein Sequence Analysis (NPSA altered in NPS@ to refer to its access by the Internet network).
 NPS@ is an interactive Web server dedicated to protein sequence analysis available for the biologist community.
NPS@ is an interactive Web server dedicated to protein sequence analysis available for the biologist community.
 NPS@ is the little web "brother" of ANTHEPROT and MPSA software. The guiding philosophy is the same for these two software and for this "webware". We hope they are useful and user-friendly.
NPS@ is the little web "brother" of ANTHEPROT and MPSA software. The guiding philosophy is the same for these two software and for this "webware". We hope they are useful and user-friendly.
 NPS@ is one service of the "Pôle Rhône-Alpes de Bio-Informatique" (PRABI).
NPS@ is one service of the "Pôle Rhône-Alpes de Bio-Informatique" (PRABI).
 NPS@'s history
NPS@'s history
 What kind of analysis can you carry out with NPS@ ?
What kind of analysis can you carry out with NPS@ ?
 What do you need to use NPS@ ?
What do you need to use NPS@ ?
 What are NPS@'s strong points ?
What are NPS@'s strong points ?
 NPS@'s flow chart.
NPS@'s flow chart.
 NPS@'s help index page.
NPS@'s help index page.
 To cite NPS@:
To cite NPS@: NPS@'s limitations:
NPS@'s limitations:
 View number of jobs performed by NPS@.
View number of jobs performed by NPS@.